b440f775ed5ac84ccb78664bce026c6a675a6af9
 ---
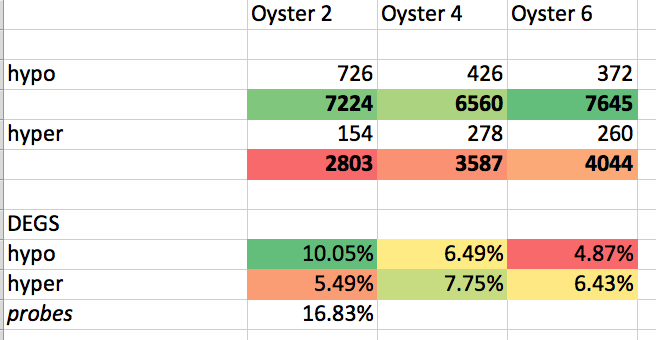
Interesting and maybe not unexpected? was that there is a clear pattern based on gene function.
---
Interesting and maybe not unexpected? was that there is a clear pattern based on gene function.
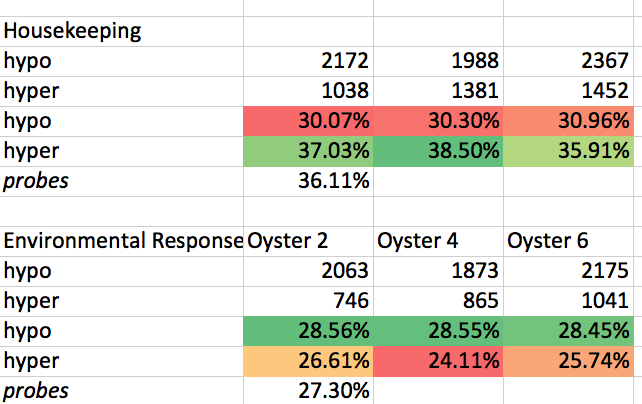 When splitting DMLs, a housekeeping genes were hyper more? hypermethylated, whereas environmental response genes were (on percentage basis) more likely to be hypomethylated. That being said it seems that the fact that both were more hypomethyated when you just look at the #s (recalling that overall there was more DMLs that were hypo v hyper.
--
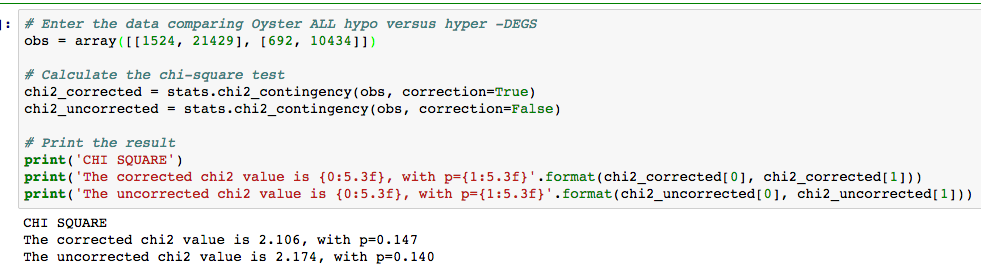
Slow story short... this is becoming a descriptive paper with clean RNA-seq data but a loss to how the array data relates. Hindsigting there are serval hypotheses.
* Stress demethylates response genes to allow for spuriousness
* Stress regulates gene expression via promoter methylation
* Stress randomly alters methylation - to add noise
* Methylation status is not impacted directly by stress but rather a side effect of gene activity.
One place I could go next is to identify gene products that significantly altered the number of isoforms post stress? This would be interesting regardless, though not readily evident how it would work .
diff --git a/notes/splicing-around.md b/notes/splicing-around.md
new file mode 100644
index 0000000..1636a5d
--- /dev/null
+++ b/notes/splicing-around.md
When splitting DMLs, a housekeeping genes were hyper more? hypermethylated, whereas environmental response genes were (on percentage basis) more likely to be hypomethylated. That being said it seems that the fact that both were more hypomethyated when you just look at the #s (recalling that overall there was more DMLs that were hypo v hyper.
--
Slow story short... this is becoming a descriptive paper with clean RNA-seq data but a loss to how the array data relates. Hindsigting there are serval hypotheses.
* Stress demethylates response genes to allow for spuriousness
* Stress regulates gene expression via promoter methylation
* Stress randomly alters methylation - to add noise
* Methylation status is not impacted directly by stress but rather a side effect of gene activity.
One place I could go next is to identify gene products that significantly altered the number of isoforms post stress? This would be interesting regardless, though not readily evident how it would work .
diff --git a/notes/splicing-around.md b/notes/splicing-around.md
new file mode 100644
index 0000000..1636a5d
--- /dev/null
+++ b/notes/splicing-around.md
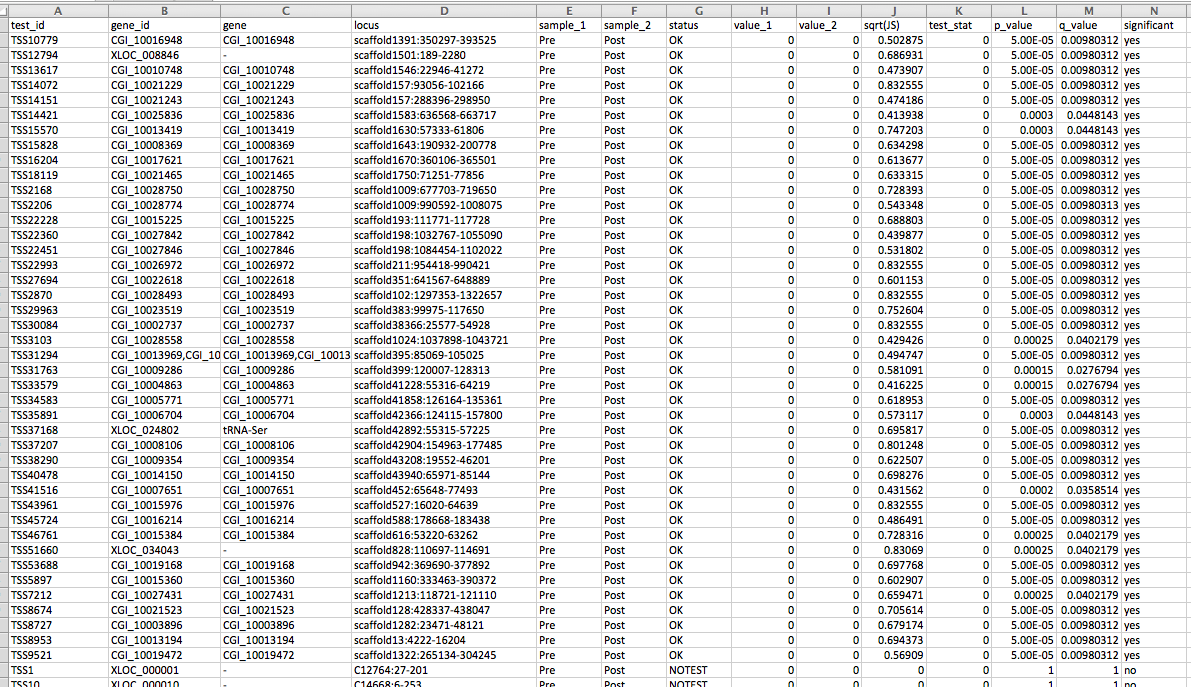
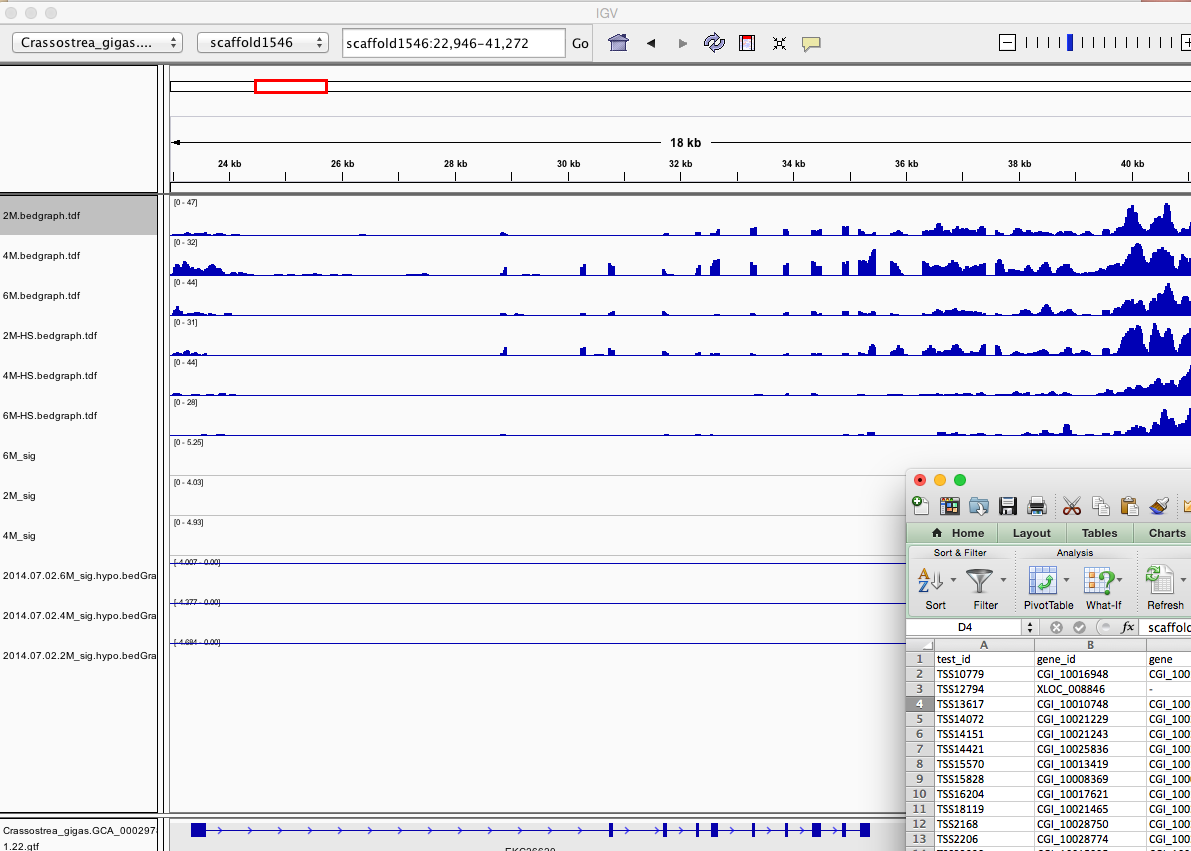
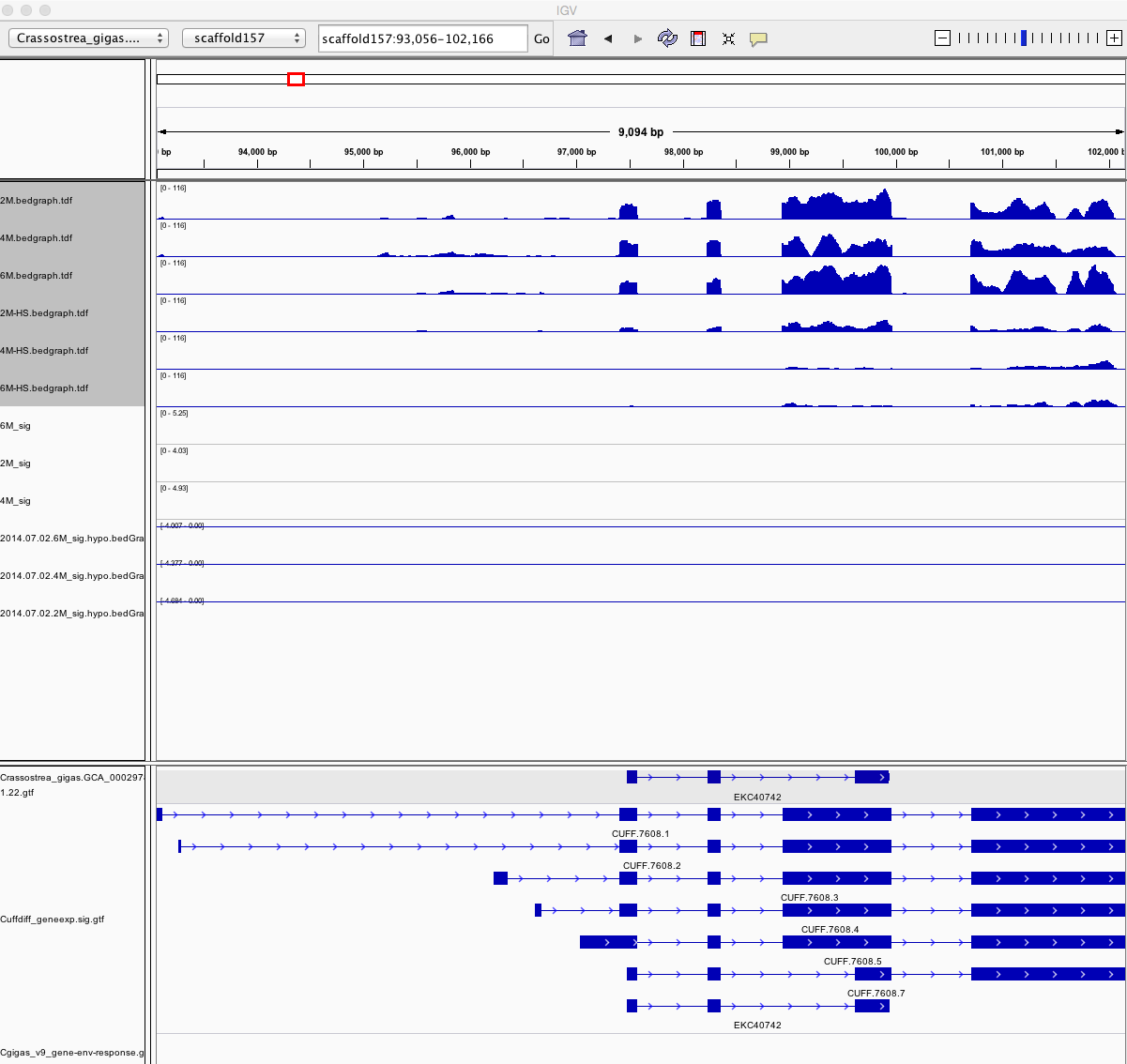
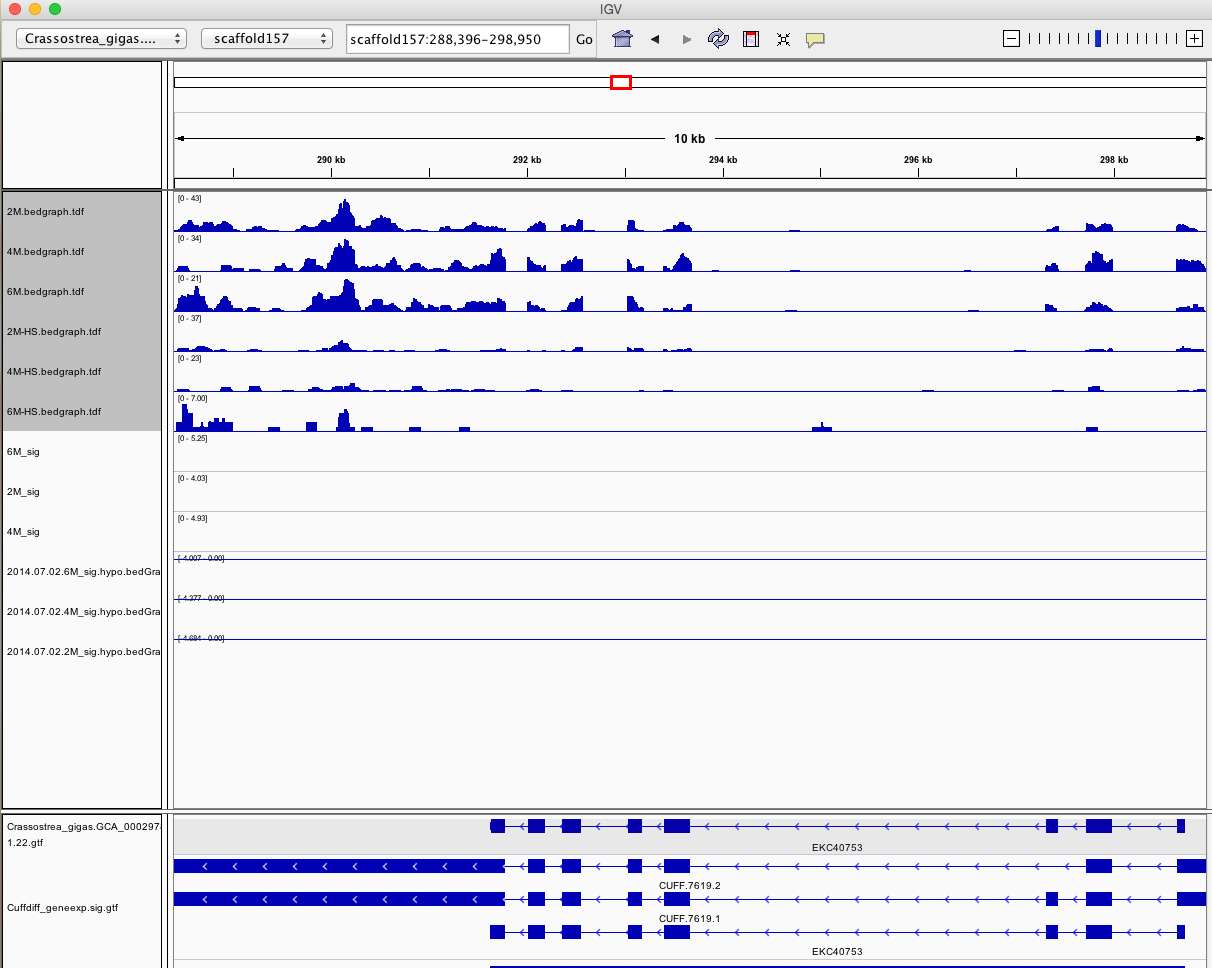
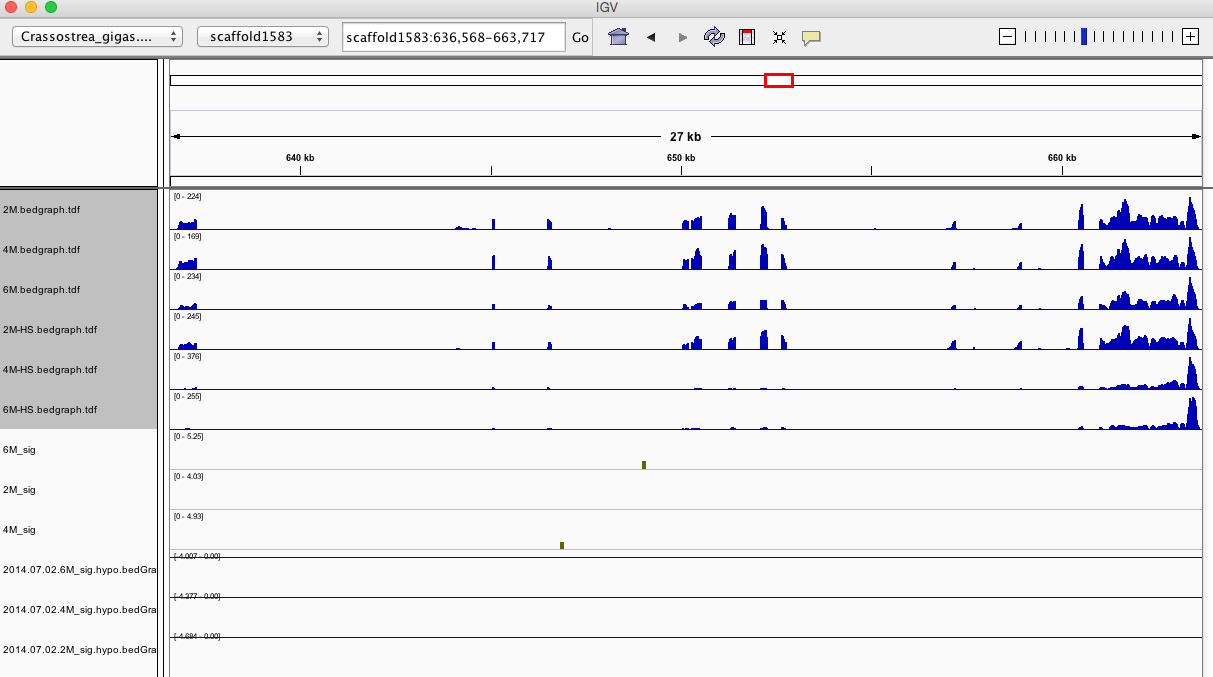
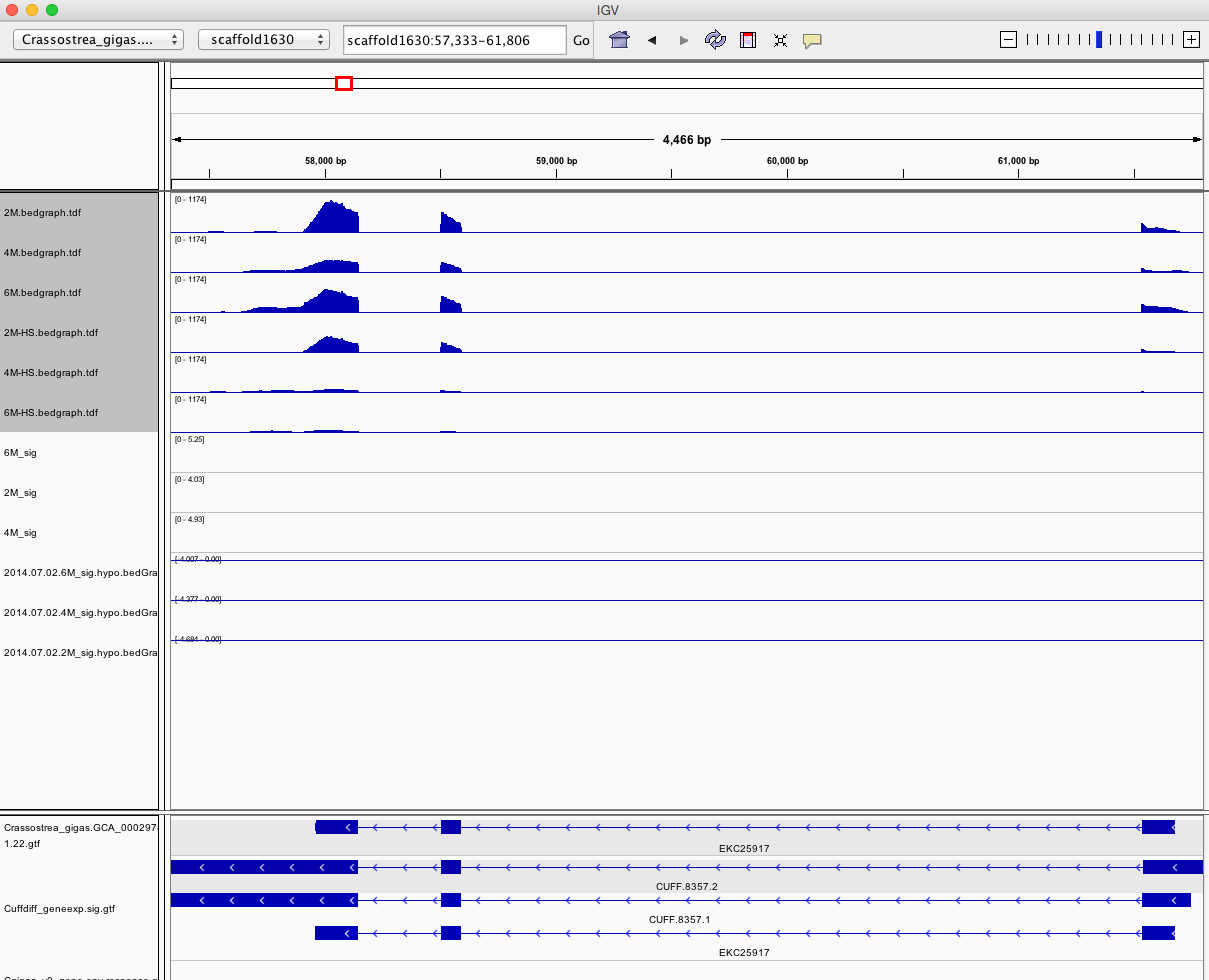
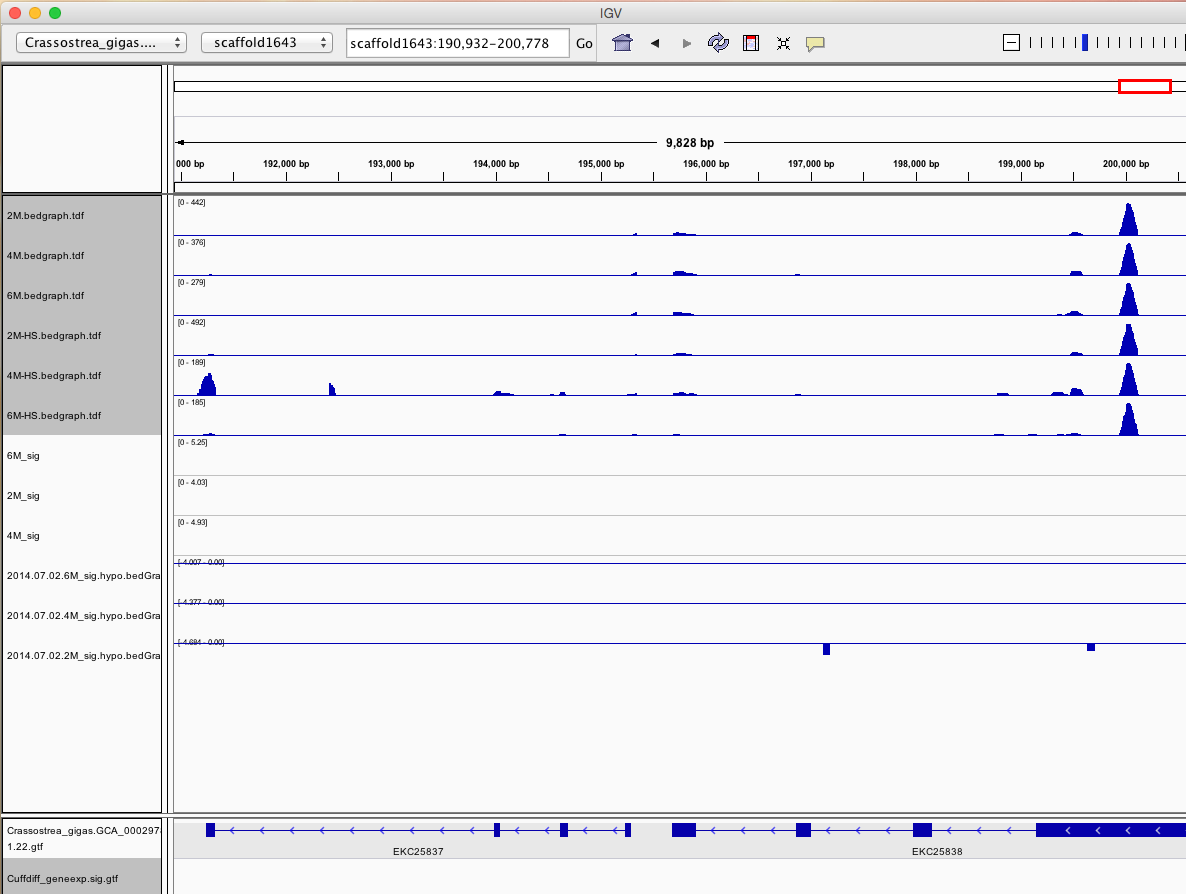
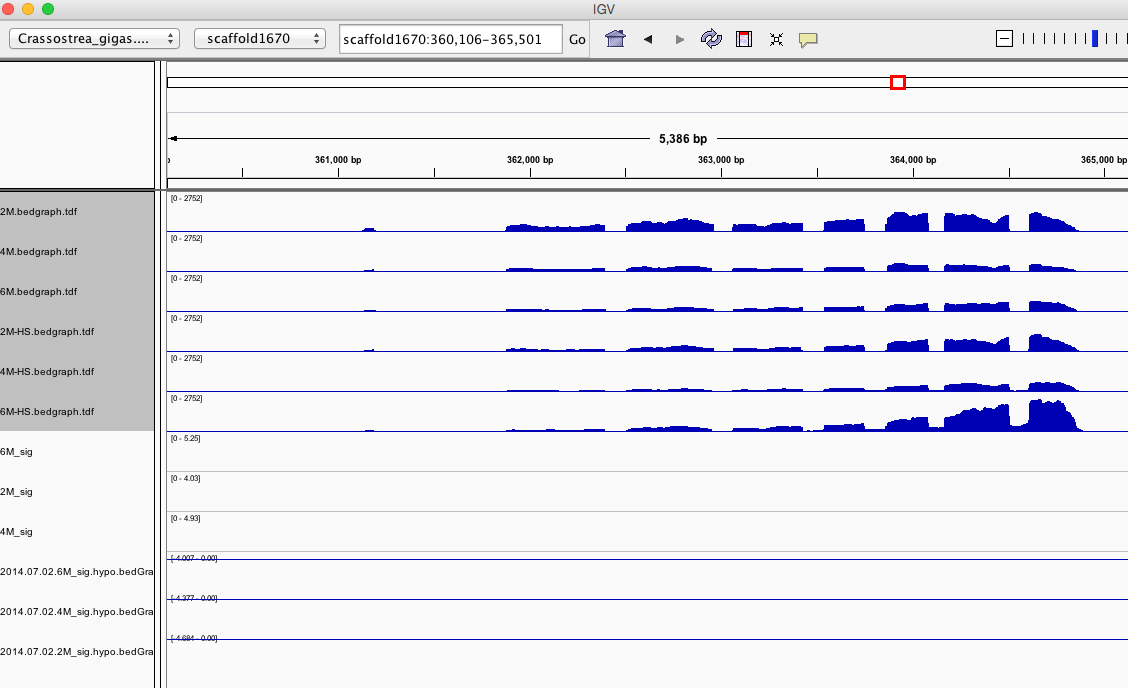
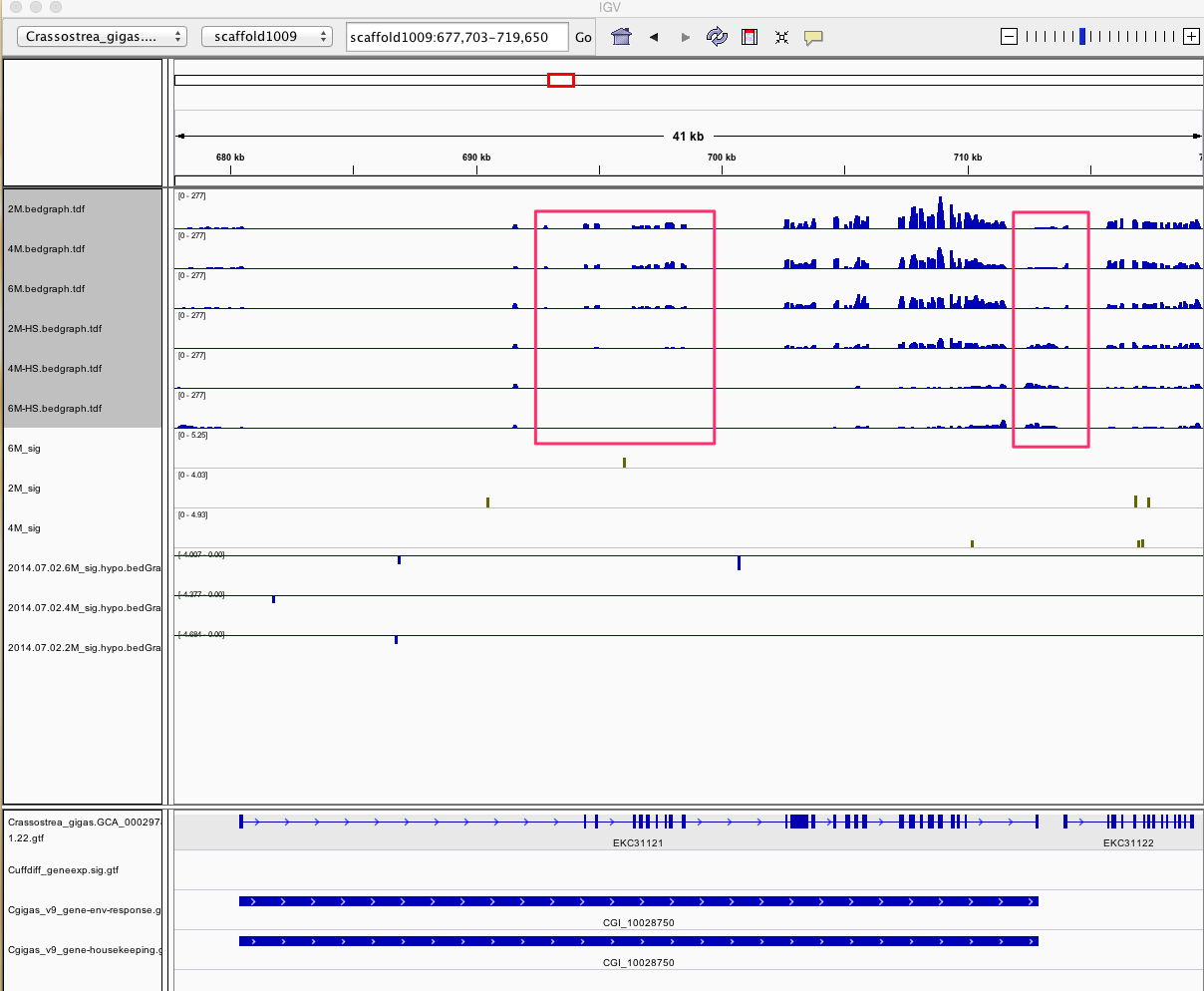
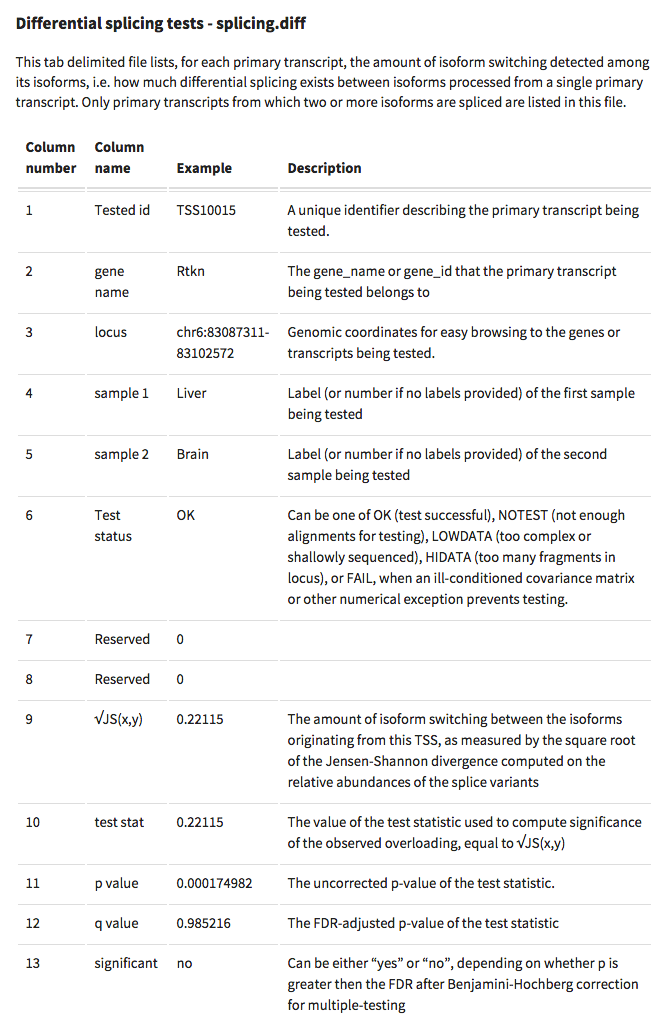 Having a gander at this file, there are in fact some features that appear to be significant. To make myself feel better about what I am looking at I will visualize in IGV.
Having a gander at this file, there are in fact some features that appear to be significant. To make myself feel better about what I am looking at I will visualize in IGV.
 ----
If my notebook holds up I should be able to refer to [this post](http://onsnetwork.org/halfshell/2015/02/26/heating-up-the-beds/) (found via IGV tag) to recreate...
----
If my notebook holds up I should be able to refer to [this post](http://onsnetwork.org/halfshell/2015/02/26/heating-up-the-beds/) (found via IGV tag) to recreate...
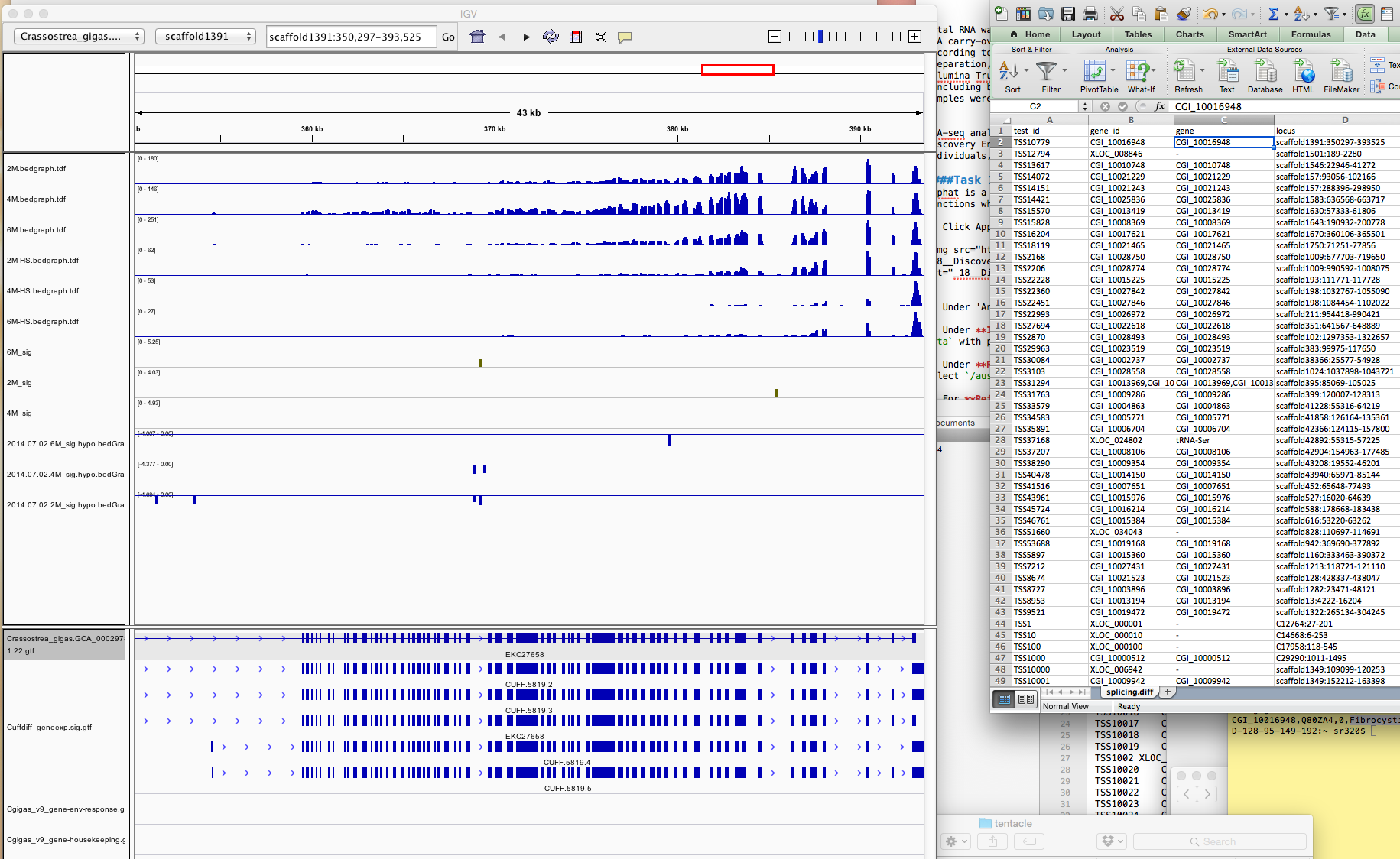
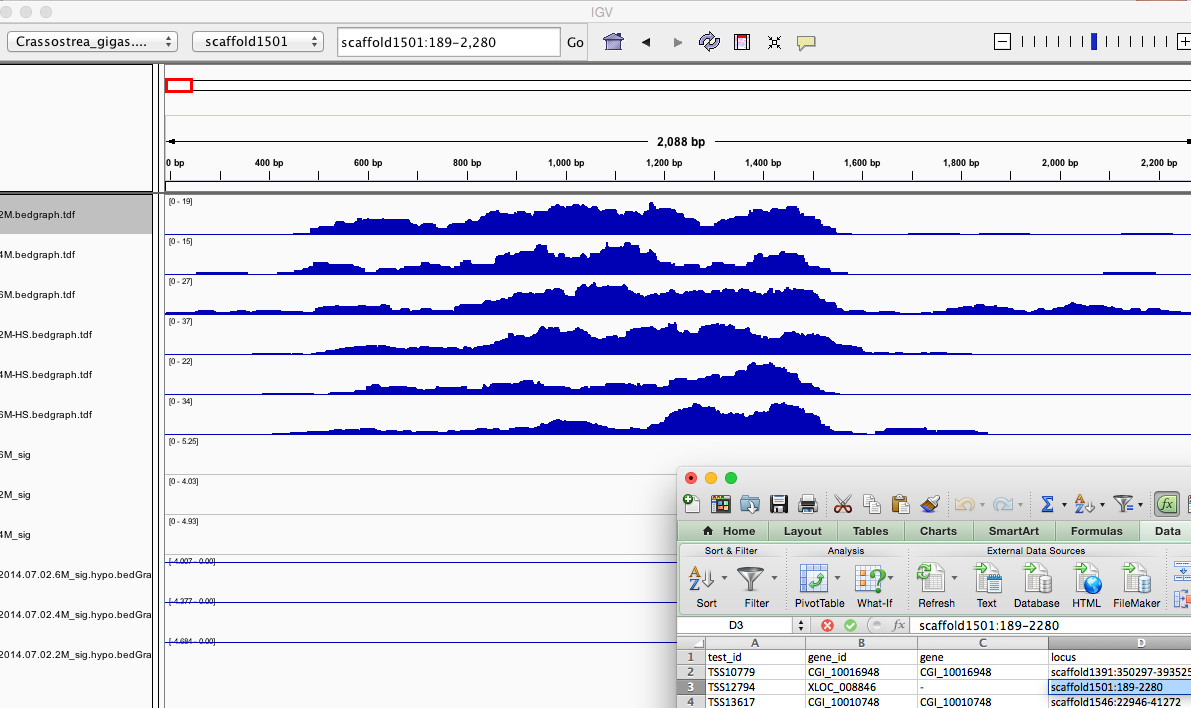
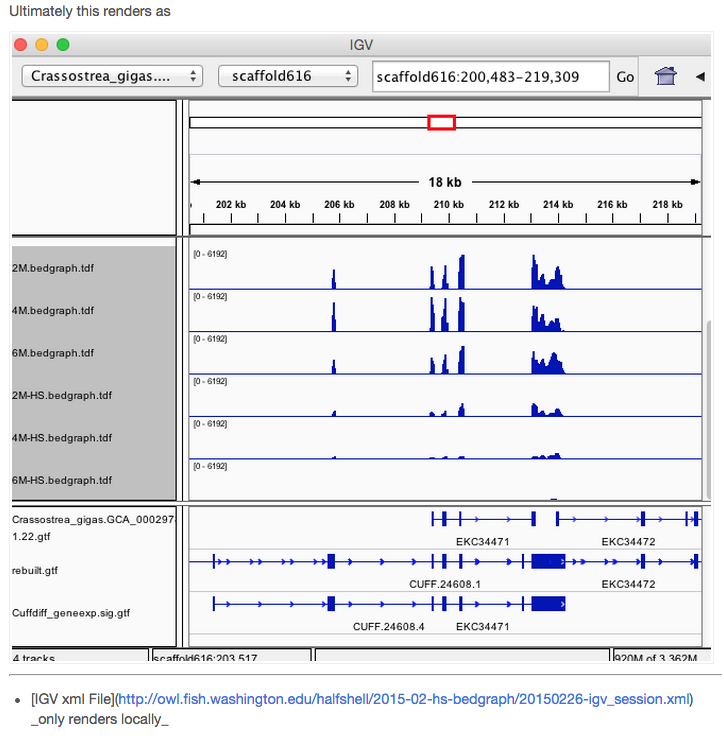 Once IGV is open I hope to simply paste locus field (ie `scaffold1391:350297-393525` into search bar. Actually there is some [fancy formulas that would allow me to directly linkout from Excel](https://www.broadinstitute.org/software/igv/ControlIGV).
----
Once IGV is open I hope to simply paste locus field (ie `scaffold1391:350297-393525` into search bar. Actually there is some [fancy formulas that would allow me to directly linkout from Excel](https://www.broadinstitute.org/software/igv/ControlIGV).
----
 ---
---
 ---
---
 ---
---
 ---
---
 ---
---
 ---
---
 ----
----
 ---
---
 ---
---
 ---
---
 ---
So there was a good chunk and cut and pastes, some things to ponder, look at, and certainly come back to.
Some closing help.
That IGV session file is @ `/Users/sr320/data-genomic/tentacle/igv_session_041615.xml`
and
Those significant splice locales
```
scaffold1391:350297-393525
scaffold1501:189-2280
scaffold1546:22946-41272
scaffold157:93056-102166
scaffold157:288396-298950
scaffold1583:636568-663717
scaffold1630:57333-61806
scaffold1643:190932-200778
scaffold1670:360106-365501
scaffold1750:71251-77856
scaffold1009:677703-719650
scaffold1009:990592-1008075
scaffold193:111771-117728
scaffold198:1032767-1055090
scaffold198:1084454-1102022
scaffold211:954418-990421
scaffold351:641567-648889
scaffold102:1297353-1322657
scaffold383:99975-117650
scaffold38366:25577-54928
scaffold1024:1037898-1043721
scaffold395:85069-105025
scaffold399:120007-128313
scaffold41228:55316-64219
scaffold41858:126164-135361
scaffold42366:124115-157800
scaffold42892:55315-57225
scaffold42904:154963-177485
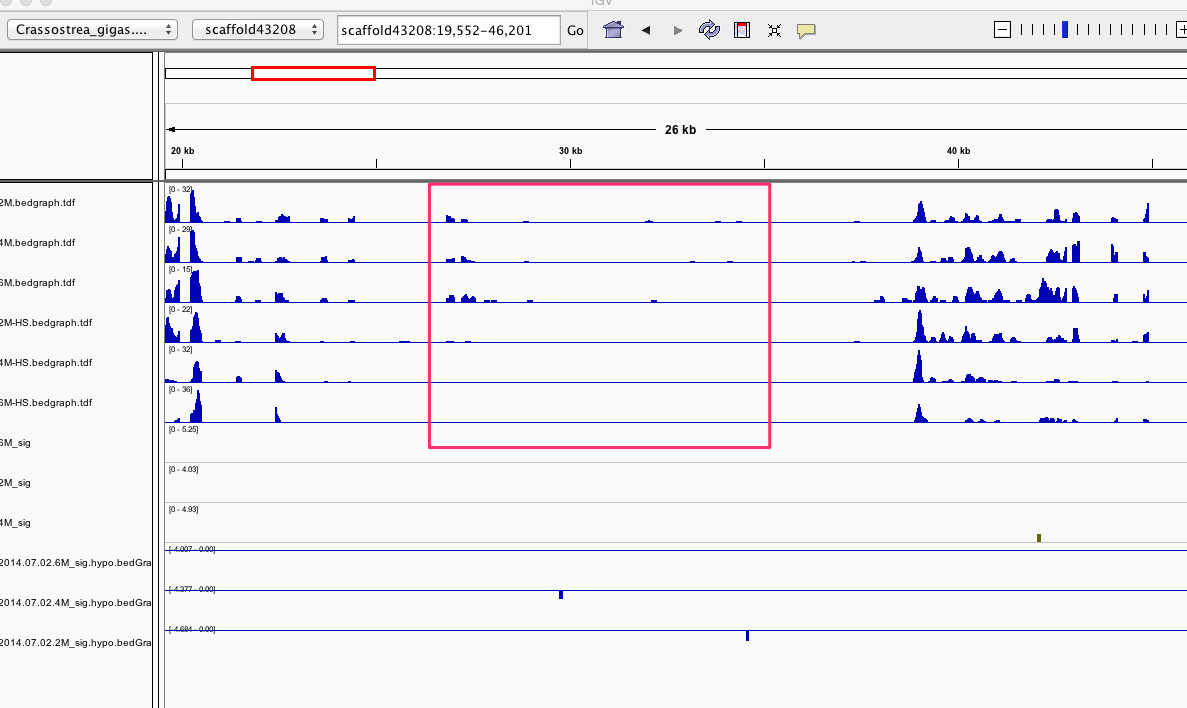
scaffold43208:19552-46201
scaffold43940:65971-85144
scaffold452:65648-77493
scaffold527:16020-64639
scaffold588:178668-183438
scaffold616:53220-63262
scaffold828:110697-114691
scaffold942:369690-377892
scaffold1160:333463-390372
scaffold1213:118721-121110
scaffold128:428337-438047
scaffold1282:23471-48121
scaffold13:4222-16204
scaffold1322:265134-304245
```
---
So there was a good chunk and cut and pastes, some things to ponder, look at, and certainly come back to.
Some closing help.
That IGV session file is @ `/Users/sr320/data-genomic/tentacle/igv_session_041615.xml`
and
Those significant splice locales
```
scaffold1391:350297-393525
scaffold1501:189-2280
scaffold1546:22946-41272
scaffold157:93056-102166
scaffold157:288396-298950
scaffold1583:636568-663717
scaffold1630:57333-61806
scaffold1643:190932-200778
scaffold1670:360106-365501
scaffold1750:71251-77856
scaffold1009:677703-719650
scaffold1009:990592-1008075
scaffold193:111771-117728
scaffold198:1032767-1055090
scaffold198:1084454-1102022
scaffold211:954418-990421
scaffold351:641567-648889
scaffold102:1297353-1322657
scaffold383:99975-117650
scaffold38366:25577-54928
scaffold1024:1037898-1043721
scaffold395:85069-105025
scaffold399:120007-128313
scaffold41228:55316-64219
scaffold41858:126164-135361
scaffold42366:124115-157800
scaffold42892:55315-57225
scaffold42904:154963-177485
scaffold43208:19552-46201
scaffold43940:65971-85144
scaffold452:65648-77493
scaffold527:16020-64639
scaffold588:178668-183438
scaffold616:53220-63262
scaffold828:110697-114691
scaffold942:369690-377892
scaffold1160:333463-390372
scaffold1213:118721-121110
scaffold128:428337-438047
scaffold1282:23471-48121
scaffold13:4222-16204
scaffold1322:265134-304245
```