this is for holding javascript data
sr320 start iplant info
over 9 years ago
Commit id: 4e584bf2761e1b5e5caa123c71adff09b16bf264
deletions | additions
diff --git a/methods-rna.md b/methods-rna.md
index 307cd33..7edd805 100644
--- a/methods-rna.md
+++ b/methods-rna.md
...
#methods-RNAseq
Total RNA was extracted using Tri-Reagent per manufacturer’s instruction. Potential DNA carry-over was removed from extracted RNA using the Turbo DNA-free treatment according to the manufacturer's instructions (Ambion). RNA quality, library preparation, and sequencing was performed by ***GENEWIZ***. Libraries were prepared using the Illumina TruSeq RNA Sample Preparation kit according to the manufacturer’s protocol (including bar-coding for multiplexing). Samples were multiplexed where three samples were run in one lane for Illumina Hiseq **100 bp paired end sequencing**.
Quality trimming of resulting sequencing reads RNA-seq analysis was
performed carried out using
CLC Genomics Workbench v. 7.0 (CLC Bio, Germany) the TopHat2 suite in the iPlant Collaborative Discovery Environment. Specfically TopHat2-SE was used to align the six libraries (3 individuals, pre and post heat shock) to the _Crassostrea gigas_ genome.
####Task 1: Align read data to _Crassostrea gigas_ genome.
Tophat is a specialized alignment software for RNA-seq reads that is aware of splice junctions when aligning to a reference assembly.
1) Click Apps from DE workspace and select **TopHat2-SE**. Use search bar.
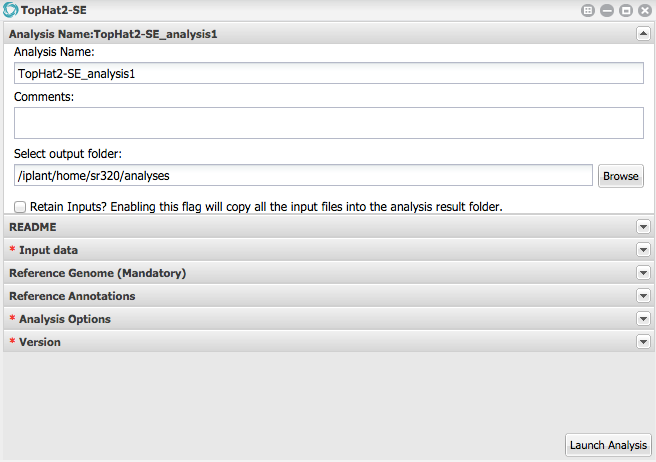 2) Under 'Analysis Name' leave as defaults for make any changes.
3) Under **Input data** for FASTQ files add six fastq.gz files localed in `austral-data`
2) Under 'Analysis Name' leave as defaults for make any changes.
3) Under **Input data** for FASTQ files add six fastq.gz files localed in `austral-data` with
prefixes _2M, 2M-HS, 4M, 4M-HS, 6M, 6M-HS_.
4) Under **Reference Genome** for 'Provide a reference genome file in FASTA format' select `/austral-data/Crassostrea_gigas.GCA_000297895.1.24.dna_sm.toplevel.fa`
5) For **Reference Annoations** add the
following parameters: quality limit = 0.05, number of ambiguous nucleotides < 2 GTF file `/austral-data/Crassostrea_gigas.GCA_000297895.1.24.gtf`
6) Click **Launch Analyses** and monitor the status of you job.
7) Once complete....
- Note this takes ~10 hours
---
####Task 2: Assemble transcripts using **Cufflinks2**
Cufflinks assembles RNA-Seq alignments into a parsimonious set of transcripts, then estimates the relative abundances of these transcripts based on
ends, how many reads
shorter support each one, taking into account biases in library preparation protocols. A detailed manual can be found at http://cufflinks.cbcb.umd.edu/manual.html.
1) Open **Cufflinks2**
2) For **Input Data** add the six bam files from the `bam` subdirectory of the TopHat2 output.
3) Under **Reference Sequence** use custom option select `/austral-data/Crassostrea_gigas.GCA_000297895.1.24.dna_sm.toplevel.fa`
4) For **Reference Annoations** add the GTF file `/austral-data/Crassostrea_gigas.GCA_000297895.1.24.gtf`
5) Click **Launch Analyses** and monitor the status of you job.
- Note this takes ~2 hours
---
####Task 3: Merge all Cufflinks transcripts into a single transcriptome annotation file using **Cuffmerge2**
Cuffmerge merges together several Cufflinks assemblies. It handles also handles running Cuffcompare. The main purpose of this application is to make it easier to make an assembly GTF file suitable for use with Cuffdiff. A merged, empirical annotation file will be more accurate than using the standard reference annotation, as the expression of rare or novel genes and alternative splicing isoforms seen in this experiment will be better reflected in the empirical transcriptome assemblies.
1) Open the **Cuffmerge2** app. Under 'Input Data', browse to the results of the cufflinks analyses (abovee) and add the 6 gtf files located in the `gtf` subdirectory.
2) For **Reference Annoations** add the GTF file `/austral-data/Crassostrea_gigas.GCA_000297895.1.24.gtf`
3) Under **Reference Sequence** use custom option select `/austral-data/Crassostrea_gigas.GCA_000297895.1.24.dna_sm.toplevel.fa`
4) Click **Launch Analyses** and monitor the status of you job.
---
####Task 4: Compare expression analysis using CuffDiff2
Cuffdiff is a program that uses the Cufflinks transcript quantification engine to calculate gene and transcript expression levels in more than
20 bp were removed, and Illumina PCR primers were removed. one condition and test them for significant differences.
1) Open **Cuffdiff2**. For 'Input Data' Sample 1 Name enter Pre adn add three bam file from Tophat analysis..
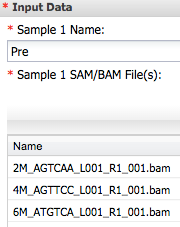 For Sample 2 enter post and add other three bam files ...
For Sample 2 enter post and add other three bam files ...
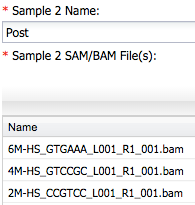 2) Next, open the **Reference Annotations** section and add a custom reference annotation file using the `merged_with_ref_ids.gtf` file from the cuffmerge output folder.
3) Click **Launch Analyses** and monitor the status of you job.
2) Next, open the **Reference Annotations** section and add a custom reference annotation file using the `merged_with_ref_ids.gtf` file from the cuffmerge output folder.
3) Click **Launch Analyses** and monitor the status of you job.
 2) Under 'Analysis Name' leave as defaults for make any changes.
3) Under **Input data** for FASTQ files add six fastq.gz files localed in `austral-data` with prefixes _2M, 2M-HS, 4M, 4M-HS, 6M, 6M-HS_.
4) Under **Reference Genome** for 'Provide a reference genome file in FASTA format' select `/austral-data/Crassostrea_gigas.GCA_000297895.1.24.dna_sm.toplevel.fa`
5) For **Reference Annoations** add the following parameters: quality limit = 0.05, number of ambiguous nucleotides < 2 GTF file `/austral-data/Crassostrea_gigas.GCA_000297895.1.24.gtf`
6) Click **Launch Analyses** and monitor the status of you job.
7) Once complete....
- Note this takes ~10 hours
---
####Task 2: Assemble transcripts using **Cufflinks2**
Cufflinks assembles RNA-Seq alignments into a parsimonious set of transcripts, then estimates the relative abundances of these transcripts based on ends, how many reads shorter support each one, taking into account biases in library preparation protocols. A detailed manual can be found at http://cufflinks.cbcb.umd.edu/manual.html.
1) Open **Cufflinks2**
2) For **Input Data** add the six bam files from the `bam` subdirectory of the TopHat2 output.
3) Under **Reference Sequence** use custom option select `/austral-data/Crassostrea_gigas.GCA_000297895.1.24.dna_sm.toplevel.fa`
4) For **Reference Annoations** add the GTF file `/austral-data/Crassostrea_gigas.GCA_000297895.1.24.gtf`
5) Click **Launch Analyses** and monitor the status of you job.
- Note this takes ~2 hours
---
####Task 3: Merge all Cufflinks transcripts into a single transcriptome annotation file using **Cuffmerge2**
Cuffmerge merges together several Cufflinks assemblies. It handles also handles running Cuffcompare. The main purpose of this application is to make it easier to make an assembly GTF file suitable for use with Cuffdiff. A merged, empirical annotation file will be more accurate than using the standard reference annotation, as the expression of rare or novel genes and alternative splicing isoforms seen in this experiment will be better reflected in the empirical transcriptome assemblies.
1) Open the **Cuffmerge2** app. Under 'Input Data', browse to the results of the cufflinks analyses (abovee) and add the 6 gtf files located in the `gtf` subdirectory.
2) For **Reference Annoations** add the GTF file `/austral-data/Crassostrea_gigas.GCA_000297895.1.24.gtf`
3) Under **Reference Sequence** use custom option select `/austral-data/Crassostrea_gigas.GCA_000297895.1.24.dna_sm.toplevel.fa`
4) Click **Launch Analyses** and monitor the status of you job.
---
####Task 4: Compare expression analysis using CuffDiff2
Cuffdiff is a program that uses the Cufflinks transcript quantification engine to calculate gene and transcript expression levels in more than 20 bp were removed, and Illumina PCR primers were removed. one condition and test them for significant differences.
1) Open **Cuffdiff2**. For 'Input Data' Sample 1 Name enter Pre adn add three bam file from Tophat analysis..
2) Under 'Analysis Name' leave as defaults for make any changes.
3) Under **Input data** for FASTQ files add six fastq.gz files localed in `austral-data` with prefixes _2M, 2M-HS, 4M, 4M-HS, 6M, 6M-HS_.
4) Under **Reference Genome** for 'Provide a reference genome file in FASTA format' select `/austral-data/Crassostrea_gigas.GCA_000297895.1.24.dna_sm.toplevel.fa`
5) For **Reference Annoations** add the following parameters: quality limit = 0.05, number of ambiguous nucleotides < 2 GTF file `/austral-data/Crassostrea_gigas.GCA_000297895.1.24.gtf`
6) Click **Launch Analyses** and monitor the status of you job.
7) Once complete....
- Note this takes ~10 hours
---
####Task 2: Assemble transcripts using **Cufflinks2**
Cufflinks assembles RNA-Seq alignments into a parsimonious set of transcripts, then estimates the relative abundances of these transcripts based on ends, how many reads shorter support each one, taking into account biases in library preparation protocols. A detailed manual can be found at http://cufflinks.cbcb.umd.edu/manual.html.
1) Open **Cufflinks2**
2) For **Input Data** add the six bam files from the `bam` subdirectory of the TopHat2 output.
3) Under **Reference Sequence** use custom option select `/austral-data/Crassostrea_gigas.GCA_000297895.1.24.dna_sm.toplevel.fa`
4) For **Reference Annoations** add the GTF file `/austral-data/Crassostrea_gigas.GCA_000297895.1.24.gtf`
5) Click **Launch Analyses** and monitor the status of you job.
- Note this takes ~2 hours
---
####Task 3: Merge all Cufflinks transcripts into a single transcriptome annotation file using **Cuffmerge2**
Cuffmerge merges together several Cufflinks assemblies. It handles also handles running Cuffcompare. The main purpose of this application is to make it easier to make an assembly GTF file suitable for use with Cuffdiff. A merged, empirical annotation file will be more accurate than using the standard reference annotation, as the expression of rare or novel genes and alternative splicing isoforms seen in this experiment will be better reflected in the empirical transcriptome assemblies.
1) Open the **Cuffmerge2** app. Under 'Input Data', browse to the results of the cufflinks analyses (abovee) and add the 6 gtf files located in the `gtf` subdirectory.
2) For **Reference Annoations** add the GTF file `/austral-data/Crassostrea_gigas.GCA_000297895.1.24.gtf`
3) Under **Reference Sequence** use custom option select `/austral-data/Crassostrea_gigas.GCA_000297895.1.24.dna_sm.toplevel.fa`
4) Click **Launch Analyses** and monitor the status of you job.
---
####Task 4: Compare expression analysis using CuffDiff2
Cuffdiff is a program that uses the Cufflinks transcript quantification engine to calculate gene and transcript expression levels in more than 20 bp were removed, and Illumina PCR primers were removed. one condition and test them for significant differences.
1) Open **Cuffdiff2**. For 'Input Data' Sample 1 Name enter Pre adn add three bam file from Tophat analysis..
 For Sample 2 enter post and add other three bam files ...
For Sample 2 enter post and add other three bam files ...
 2) Next, open the **Reference Annotations** section and add a custom reference annotation file using the `merged_with_ref_ids.gtf` file from the cuffmerge output folder.
3) Click **Launch Analyses** and monitor the status of you job.
2) Next, open the **Reference Annotations** section and add a custom reference annotation file using the `merged_with_ref_ids.gtf` file from the cuffmerge output folder.
3) Click **Launch Analyses** and monitor the status of you job.